You are using an unsupported browser. Please upgrade your browser to a newer version to get the best experience on Toxin, Toxin Target Database.
Morphine (T3D2740)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2009-07-21 20:26:30 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:25:51 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D2740 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Morphine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Morphine is only found in individuals that have used or taken this drug. It is the principal alkaloid in opium and the prototype opiate analgesic and narcotic. Morphine has widespread effects in the central nervous system and on smooth muscle. The precise mechanism of the analgesic action of morphine is unknown. However, specific CNS opiate receptors have been identified and likely play a role in the expression of analgesic effects. Morphine first acts on the mu-opioid receptors. The mechanism of respiratory depression involves a reduction in the responsiveness of the brain stem respiratory centers to increases in carbon dioxide tension and to electrical stimulation.It has been shown that morphine binds to and inhibits GABA inhibitory interneurons. These interneurons normally inhibit the descending pain inhibition pathway. So, without the inhibitory signals, pain modulation can proceed downstream. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
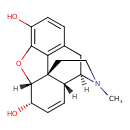
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C17H19NO3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 285.338 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 285.136 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 57-27-2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | (1S,5R,13R,14S,17R)-4-methyl-12-oxa-4-azapentacyclo[9.6.1.0¹,¹³.0⁵,¹⁷.0⁷,¹⁸]octadeca-7(18),8,10,15-tetraene-10,14-diol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | morph | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [H][C@@]12OC3=C(O)C=CC4=C3[C@@]11CCN(C)[C@]([H])(C4)[C@]1([H])C=C[C@]2([H])O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C17H19NO3/c1-18-7-6-17-10-3-5-13(20)16(17)21-15-12(19)4-2-9(14(15)17)8-11(10)18/h2-5,10-11,13,16,19-20H,6-8H2,1H3/t10-,11+,13-,16-,17-/m0/s1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=BQJCRHHNABKAKU-KBQPJGBKSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as morphinans. These are polycyclic compounds with a four-ring skeleton with three condensed six-member rings forming a partially hydrogenated phenanthrene moiety, one of which is aromatic while the two others are alicyclic. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Alkaloids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Morphinans | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Morphinans | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Intravenous, Topical, Oral, Intramuscular, Epidural, Rectal. Bioavailability is approximately 30%. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | The precise mechanism of the analgesic action of morphine is unknown. However, specific CNS opiate receptors have been identified and likely play a role in the expression of analgesic effects. Morphine first acts on the mu-opioid receptors. The mechanism of respiratory depression involves a reduction in the responsiveness of the brain stem respiratory centers to increases in carbon dioxide tension and to electrical stimulation. It has been shown that morphine binds to and inhibits GABA inhibitory interneurons. These interneurons normally inhibit the descending pain inhibition pathway. So, without the inhibitory signals, pain modulation can proceed downstream. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Primarily hepatic (90%), converted to dihydromorphinone and normorphine. Also converted to morphine-3-glucuronide (M3G) and morphine-6-glucuronide. Virtually all morphine is converted to glucuronide metabolites; only a small fraction (less than 5%) of absorbed morphine is demethylated. Route of Elimination: A small amount of glucuronide conjugates are excreted in bile, with minor enterohepatic recycling. Seven to 10% of administered morphine sulfate is excreted in the feces. Half Life: 2-4 hours | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | LD50: 461 mg/kg (oral, rat) LD50: 600 mg/kg (oral, mouse) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Human lethal dose by ingestion is 120-250 mg of morphine sulfate. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | For the relief and treatment of severe pain. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Medical problems can include congested lungs, liver disease, tetanus, infection of the heart valves, skin abscesses, anemia and pneumonia. Death can occur from overdose. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Symptoms of overdose include cold, clammy skin, flaccid muscles, fluid in the lungs, lowered blood pressure, "pinpoint" or dilated pupils, sleepiness leading to stupor and coma, slowed breathing, and slow pulse rate. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | In the treatment of morphine overdosage, primary attention should be given to the re- establishment of a patent airway and institution of assisted or controlled ventilation. Supportive measures (including oxygen, vasopressors) should be employed in the management of circulatory shock and pulmonary edema accompanying overdose as indicated. Cardiac arrest or arrhythmias may require cardiac massage or defibrillation. The pure opioid antagonists, such as naloxone, are specific antidotes against respiratory depression which results from opioid overdose. Naloxone should be administered intravenously; however, because its duration of action is relatively short, the patient must be carefully monitored until spontaneous respiration is reliably re-established. If the response to naloxone is suboptimal or not sustained, additional naloxone may be administered, as needed, or given by continuous infusion to maintain alertness and respiratory function; however, there is no information available about the cumulative dose of naloxone that may be safely administered. (4) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | DB00295 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB14440 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 5288826 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL70 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 4450907 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C01516 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 17303 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | MORPHINE | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Morphine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | MOI | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Morphine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Nagaraj R. Ayyangar, Anil R. Choudhary, Uttam R. Kalkote, Vasant K. Sharma, “Process for the preparation of codeine from morphine.” U.S. Patent US4764615, issued May, 1912. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Link | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Opioid receptor activity
- Specific Function:
- G-protein coupled opioid receptor that functions as receptor for endogenous alpha-neoendorphins and dynorphins, but has low affinity for beta-endorphins. Also functions as receptor for various synthetic opioids and for the psychoactive diterpene salvinorin A. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors, such as adenylate cyclase. Signaling leads to the inhibition of adenylate cyclase activity. Inhibits neurotransmitter release by reducing calcium ion currents and increasing potassium ion conductance. Plays a role in the perception of pain. Plays a role in mediating reduced physical activity upon treatment with synthetic opioids. Plays a role in the regulation of salivation in response to synthetic opioids. May play a role in arousal and regulation of autonomic and neuroendocrine functions.
- Gene Name:
- OPRK1
- Uniprot ID:
- P41145
- Molecular Weight:
- 42644.665 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 0.0069 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.0111 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.0133 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.024 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.033 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.0337 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.0469 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.049 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.061 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.064 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.069 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.086 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.11 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.113 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.151 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.167 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.188 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.26 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.299 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.301 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.33 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.538 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 1.87 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 6.424 uM | Not Available | BindingDB 50000092 |
| IC50 | 0.0018 uM | Not Available | BindingDB 50000092 |
| IC50 | 0.13 uM | Not Available | BindingDB 50000092 |
| IC50 | 0.17 uM | Not Available | BindingDB 50000092 |
| IC50 | 0.271 uM | Not Available | BindingDB 50000092 |
| IC50 | 0.53 uM | Not Available | BindingDB 50000092 |
| IC50 | 2.39 uM | Not Available | BindingDB 50000092 |
References
- Overington JP, Al-Lazikani B, Hopkins AL: How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. [17139284 ]
- Imming P, Sinning C, Meyer A: Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006 Oct;5(10):821-34. [17016423 ]
- Leventhal L, Silva RM, Rossi GC, Pasternak GW, Bodnar RJ: Morphine-6beta-glucuronide-induced hyperphagia: characterization of opioid action by selective antagonists and antisense mapping in rats. J Pharmacol Exp Ther. 1998 Nov;287(2):538-44. [9808678 ]
- Kozak CA, Filie J, Adamson MC, Chen Y, Yu L: Murine chromosomal location of the mu and kappa opioid receptor genes. Genomics. 1994 Jun;21(3):659-61. [7959748 ]
- Teodorov E, Modena CC, Sukikara MH, Felicio LF: Preliminary study of the effects of morphine treatment on opioid receptor gene expression in brain structures of the female rat. Neuroscience. 2006 Sep 1;141(3):1225-31. Epub 2006 Jun 6. [16753266 ]
- Bertha CM, Flippen-Anderson JL, Rothman RB, Porreca F, Davis P, Xu H, Becketts K, Cha XY, Rice KC: Probes for narcotic receptor-mediated phenomena. 20. Alteration of opioid receptor subtype selectivity of the 5-(3-hydroxyphenyl)morphans by application of the message-address concept: preparation of delta-opioid receptor ligands. J Med Chem. 1995 Apr 28;38(9):1523-37. [7739011 ]
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK: BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007 Jan;35(Database issue):D198-201. Epub 2006 Dec 1. [17145705 ]
- Hanessian S, Parthasarathy S, Mauduit M, Payza K: The power of visual imagery in drug design. Isopavines as a new class of morphinomimetics and their human opioid receptor binding activity. J Med Chem. 2003 Jan 2;46(1):34-48. [12502358 ]
- Bertha CM, Ellis M, Flippen-Anderson JL, Porreca F, Rothman RB, Davis P, Xu H, Becketts K, Rice KC: Probes for narcotic receptor-mediated phenomena. 21. Novel derivatives of 3-(1,2,3,4,5,11-hexahydro-3-methyl-2,6-methano-6H-azocino[4,5-b]indol- 6-yl)-phenols with improved delta opioid receptor selectivity. J Med Chem. 1996 May 10;39(10):2081-6. [8642567 ]
- Marastoni M, Salvadori S, Balboni G, Borea PA, Marzola G, Tomatis R: Synthesis and activity profiles of new dermorphin-(1-4) peptide analogues. J Med Chem. 1987 Sep;30(9):1538-42. [2887656 ]
- Barlow JJ, Blackburn TP, Costello GF, James R, Le Count DJ, Main BG, Pearce RJ, Russell K, Shaw JS: Structure/activity studies related to 2-(3,4-dichlorophenyl)-N-methyl-N-[2-(1-pyrrolidinyl)-1-substituted- ethyl]acetamides: a novel series of potent and selective kappa-opioid agonists. J Med Chem. 1991 Nov;34(11):3149-58. [1659636 ]
- Costello GF, James R, Shaw JS, Slater AM, Stutchbury NC: 2-(3,4-Dichlorophenyl)-N-methyl-N-[2-(1-pyrrolidinyl)-1-substituted- ethyl]-acetamides: the use of conformational analysis in the development of a novel series of potent opioid kappa agonists. J Med Chem. 1991 Jan;34(1):181-9. [1846918 ]
- Li W, Tao YM, Tang Y, Xu XJ, Chen J, Fu W, Wang XH, Chao B, Sheng W, Xie Q, Qiu ZB, Liu JG: Highly selective and potent mu opioid ligands by unexpected substituent on morphine skeleton. Bioorg Med Chem Lett. 2010 Jan 1;20(1):418-21. doi: 10.1016/j.bmcl.2009.07.119. Epub 2009 Jul 29. [19932964 ]
- Standifer KM, Cheng J, Brooks AI, Honrado CP, Su W, Visconti LM, Biedler JL, Pasternak GW: Biochemical and pharmacological characterization of mu, delta and kappa 3 opioid receptors expressed in BE(2)-C neuroblastoma cells. J Pharmacol Exp Ther. 1994 Sep;270(3):1246-55. [7932177 ]
- Varamini P, Mansfeld FM, Blanchfield JT, Wyse BD, Smith MT, Toth I: Synthesis and biological evaluation of an orally active glycosylated endomorphin-1. J Med Chem. 2012 Jun 28;55(12):5859-67. doi: 10.1021/jm300418d. Epub 2012 Jun 15. [22680612 ]
- Wentland MP, Duan W, Cohen DJ, Bidlack JM: Selective protection and functionalization of morphine: synthesis and opioid receptor binding properties of 3-amino-3-desoxymorphine derivatives. J Med Chem. 2000 Sep 21;43(19):3558-65. [11000010 ]
- Zhang A, Xiong W, Hilbert JE, DeVita EK, Bidlack JM, Neumeyer JL: 2-aminothiazole-derived opioids. Bioisosteric replacement of phenols. J Med Chem. 2004 Apr 8;47(8):1886-8. [15055988 ]
- Peng X, Knapp BI, Bidlack JM, Neumeyer JL: Synthesis and preliminary in vitro investigation of bivalent ligands containing homo- and heterodimeric pharmacophores at mu, delta, and kappa opioid receptors. J Med Chem. 2006 Jan 12;49(1):256-62. [16392810 ]
- Neumeyer JL, Zhang B, Zhang T, Sromek AW, Knapp BI, Cohen DJ, Bidlack JM: Synthesis, binding affinity, and functional in vitro activity of 3-benzylaminomorphinan and 3-benzylaminomorphine ligands at opioid receptors. J Med Chem. 2012 Apr 26;55(8):3878-90. doi: 10.1021/jm3001086. Epub 2012 Apr 4. [22439881 ]
- Froimowitz M, Pick CG, Pasternak GW: Phenylmorphans and analogues: opioid receptor subtype selectivity and effect of conformation on activity. J Med Chem. 1992 May 1;35(9):1521-5. [1315868 ]
- Yamaotsu N, Fujii H, Nagase H, Hirono S: Identification of the three-dimensional pharmacophore of kappa-opioid receptor agonists. Bioorg Med Chem. 2010 Jun 15;18(12):4446-52. doi: 10.1016/j.bmc.2010.04.069. Epub 2010 Apr 28. [20478711 ]
- Toll L, Berzetei-Gurske IP, Polgar WE, Brandt SR, Adapa ID, Rodriguez L, Schwartz RW, Haggart D, O'Brien A, White A, Kennedy JM, Craymer K, Farrington L, Auh JS: Standard binding and functional assays related to medications development division testing for potential cocaine and opiate narcotic treatment medications. NIDA Res Monogr. 1998 Mar;178:440-66. [9686407 ]
- Nieland NP, Moynihan HA, Carrington S, Broadbear J, Woods JH, Traynor JR, Husbands SM, Lewis JW: Structural determinants of opioid activity in derivatives of 14-aminomorphinones: effect of substitution in the aromatic ring of cinnamoylaminomorphinones and codeinones. J Med Chem. 2006 Aug 24;49(17):5333-8. [16913723 ]
- Rennison D, Neal AP, Cami-Kobeci G, Aceto MD, Martinez-Bermejo F, Lewis JW, Husbands SM: Cinnamoyl derivatives of 7alpha-aminomethyl-6,14-endo-ethanotetrahydrothebaine and 7alpha-aminomethyl-6,14-endo-ethanotetrahydrooripavine and related opioid ligands. J Med Chem. 2007 Oct 18;50(21):5176-82. Epub 2007 Sep 22. [17887741 ]
- Mosberg HI, Yeomans L, Harland AA, Bender AM, Sobczyk-Kojiro K, Anand JP, Clark MJ, Jutkiewicz EM, Traynor JR: Opioid peptidomimetics: leads for the design of bioavailable mixed efficacy mu opioid receptor (MOR) agonist/delta opioid receptor (DOR) antagonist ligands. J Med Chem. 2013 Mar 14;56(5):2139-49. doi: 10.1021/jm400050y. Epub 2013 Feb 27. [23419026 ]
- Anzini M, Canullo L, Braile C, Cappelli A, Gallelli A, Vomero S, Menziani MC, De Benedetti PG, Rizzo M, Collina S, Azzolina O, Sbacchi M, Ghelardini C, Galeotti N: Synthesis, biological evaluation, and receptor docking simulations of 2-[(acylamino)ethyl]-1,4-benzodiazepines as kappa-opioid receptor agonists endowed with antinociceptive and antiamnesic activity. J Med Chem. 2003 Aug 28;46(18):3853-64. [12930147 ]
- Brine GA, Stark PA, Liu Y, Carroll FI, Singh P, Xu H, Rothman RB: Enantiomers of diastereomeric cis-N-[1-(2-hydroxy-2-phenylethyl)- 3-methyl-4-piperidyl]-N-phenylpropanamides: synthesis, X-ray analysis, and biological activities. J Med Chem. 1995 Apr 28;38(9):1547-57. [7739013 ]
- Halfpenny PR, Hill RG, Horwell DC, Hughes J, Hunter JC, Johnson S, Rees DC: Highly selective kappa-opioid analgesics. 2. Synthesis and structure-activity relationships of novel N-[(2-aminocyclohexyl)aryl]acetamide derivatives. J Med Chem. 1989 Jul;32(7):1620-6. [2567782 ]
- Clark CR, Halfpenny PR, Hill RG, Horwell DC, Hughes J, Jarvis TC, Rees DC, Schofield D: Highly selective kappa opioid analgesics. Synthesis and structure-activity relationships of novel N-[(2-aminocyclohexyl)aryl]acetamide and N-[(2-aminocyclohexyl)aryloxy]acetamide derivatives. J Med Chem. 1988 Apr;31(4):831-6. [2832603 ]
- Wang C, McFadyen IJ, Traynor JR, Mosberg HI: Design of a high affinity peptidomimetic opioid agonist from peptide pharmacophore models. Bioorg Med Chem Lett. 1998 Oct 6;8(19):2685-8. [9873603 ]
- Greiner E, Spetea M, Krassnig R, Schullner F, Aceto M, Harris LS, Traynor JR, Woods JH, Coop A, Schmidhammer H: Synthesis and biological evaluation of 14-alkoxymorphinans. 18. N-substituted 14-phenylpropyloxymorphinan-6-ones with unanticipated agonist properties: extending the scope of common structure-activity relationships. J Med Chem. 2003 Apr 24;46(9):1758-63. [12699394 ]
- Vecchietti V, Giordani A, Giardina G, Colle R, Clarke GD: (2S)-1-(arylacetyl)-2-(aminomethyl)piperidine derivatives: novel, highly selective kappa opioid analgesics. J Med Chem. 1991 Jan;34(1):397-403. [1846921 ]
- Zimmerman DM, Cantrell BE, Swartzendruber JK, Jones ND, Mendelsohn LG, Leander JD, Nickander RC: Synthesis and analgesic properties of N-substituted trans-4a-aryldecahydroisoquinolines. J Med Chem. 1988 Mar;31(3):555-60. [2831363 ]
- Ananthan S, Khare NK, Saini SK, Seitz LE, Bartlett JL, Davis P, Dersch CM, Porreca F, Rothman RB, Bilsky EJ: Identification of opioid ligands possessing mixed micro agonist/delta antagonist activity among pyridomorphinans derived from naloxone, oxymorphone, and hydromorphone [correction of hydropmorphone]. J Med Chem. 2004 Mar 11;47(6):1400-12. [14998329 ]
- Shreder K, Zhang L, Dang T, Yaksh TL, Umeno H, DeHaven R, Daubert J, Goodman M: Synthesis and biological activity of a novel methylamine-bridged enkephalin analogue (MABE): a new route to cyclic peptides and peptidomimetics. J Med Chem. 1998 Jul 2;41(14):2631-5. [9651168 ]
- Rew Y, Malkmus S, Svensson C, Yaksh TL, Chung NN, Schiller PW, Cassel JA, DeHaven RN, Taulane JP, Goodman M: Synthesis and biological activities of cyclic lanthionine enkephalin analogues: delta-opioid receptor selective ligands. J Med Chem. 2002 Aug 15;45(17):3746-54. [12166947 ]
- Iyer MR, Lee YS, Deschamps JR, Dersch CM, Rothman RB, Jacobson AE, Rice KC: Probes for narcotic receptor mediated phenomena. 44. Synthesis of an N-substituted 4-hydroxy-5-(3-hydroxyphenyl)morphan with high affinity and selective mu-antagonist activity. Eur J Med Chem. 2012 Apr;50:44-54. doi: 10.1016/j.ejmech.2012.01.025. Epub 2012 Jan 20. [22341895 ]
- Vecchietti V, Clarke GD, Colle R, Dondio G, Giardina G, Petrone G, Sbacchi M: Substituted 1-(aminomethyl)-2-(arylacetyl)-1,2,3,4-tetrahydroisoquinolines: a novel class of very potent antinociceptive agents with varying degrees of selectivity for kappa and mu opioid receptors. J Med Chem. 1992 Aug 7;35(16):2970-8. [1323679 ]
- Vecchietti V, Clarke GD, Colle R, Giardina G, Petrone G, Sbacchi M: (1S)-1-(aminomethyl)-2-(arylacetyl)-1,2,3,4-tetrahydroisoquinoline and heterocycle-condensed tetrahydropyridine derivatives: members of a novel class of very potent kappa opioid analgesics. J Med Chem. 1991 Aug;34(8):2624-33. [1652025 ]
- Schultz AG, Wang A, Alva C, Sebastian A, Glick SD, Deecher DC, Bidlack JM: Asymmetric syntheses, opioid receptor affinities, and antinociceptive effects of 8-amino-5,9-methanobenzocyclooctenes, a new class of structural analogues of the morphine alkaloids. J Med Chem. 1996 May 10;39(10):1956-66. [8642554 ]
- Raynor K, Kong H, Chen Y, Yasuda K, Yu L, Bell GI, Reisine T: Pharmacological characterization of the cloned kappa-, delta-, and mu-opioid receptors. Mol Pharmacol. 1994 Feb;45(2):330-4. [8114680 ]
- Tsai YC, Liou JP, Liao R, Cheng CY, Tao PL: C-alkylated spiro[benzofuran-3(2H),4'-1'-methyl-piperidine-7-ols] as potent opioids: a conformation-activity study. Bioorg Med Chem Lett. 1998 Jul 21;8(14):1813-8. [9873439 ]
- Ronsisvalle G, Pasquinucci L, Pappalardo MS, Vittorio F, Fronza G, Romagnoli C, Pistacchio E, Spampinato S, Ferri S: Non-peptide ligands for opioid receptors. Design of kappa-specific agonists. J Med Chem. 1993 Jun 25;36(13):1860-5. [8390575 ]
- General Function:
- Voltage-gated calcium channel activity
- Specific Function:
- Receptor for endogenous opioids such as beta-endorphin and endomorphin. Receptor for natural and synthetic opioids including morphine, heroin, DAMGO, fentanyl, etorphine, buprenorphin and methadone. Agonist binding to the receptor induces coupling to an inactive GDP-bound heterotrimeric G-protein complex and subsequent exchange of GDP for GTP in the G-protein alpha subunit leading to dissociation of the G-protein complex with the free GTP-bound G-protein alpha and the G-protein beta-gamma dimer activating downstream cellular effectors. The agonist- and cell type-specific activity is predominantly coupled to pertussis toxin-sensitive G(i) and G(o) G alpha proteins, GNAI1, GNAI2, GNAI3 and GNAO1 isoforms Alpha-1 and Alpha-2, and to a lesser extend to pertussis toxin-insensitive G alpha proteins GNAZ and GNA15. They mediate an array of downstream cellular responses, including inhibition of adenylate cyclase activity and both N-type and L-type calcium channels, activation of inward rectifying potassium channels, mitogen-activated protein kinase (MAPK), phospholipase C (PLC), phosphoinositide/protein kinase (PKC), phosphoinositide 3-kinase (PI3K) and regulation of NF-kappa-B. Also couples to adenylate cyclase stimulatory G alpha proteins. The selective temporal coupling to G-proteins and subsequent signaling can be regulated by RGSZ proteins, such as RGS9, RGS17 and RGS4. Phosphorylation by members of the GPRK subfamily of Ser/Thr protein kinases and association with beta-arrestins is involved in short-term receptor desensitization. Beta-arrestins associate with the GPRK-phosphorylated receptor and uncouple it from the G-protein thus terminating signal transduction. The phosphorylated receptor is internalized through endocytosis via clathrin-coated pits which involves beta-arrestins. The activation of the ERK pathway occurs either in a G-protein-dependent or a beta-arrestin-dependent manner and is regulated by agonist-specific receptor phosphorylation. Acts as a class A G-protein coupled receptor (GPCR) which dissociates from beta-arrestin at or near the plasma membrane and undergoes rapid recycling. Receptor down-regulation pathways are varying with the agonist and occur dependent or independent of G-protein coupling. Endogenous ligands induce rapid desensitization, endocytosis and recycling whereas morphine induces only low desensitization and endocytosis. Heterooligomerization with other GPCRs can modulate agonist binding, signaling and trafficking properties. Involved in neurogenesis. Isoform 12 couples to GNAS and is proposed to be involved in excitatory effects. Isoform 16 and isoform 17 do not bind agonists but may act through oligomerization with binding-competent OPRM1 isoforms and reduce their ligand binding activity.
- Gene Name:
- OPRM1
- Uniprot ID:
- P35372
- Molecular Weight:
- 44778.855 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 0.00014 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.00088 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.0011 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.0018 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.00192 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.00252 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.00255 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.0026 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.003 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.0033 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.0046 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.00651 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.00771 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.017 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.038 uM | Not Available | BindingDB 50000092 |
| IC50 | 0.00057 uM | Not Available | BindingDB 50000092 |
| IC50 | 0.004 uM | Not Available | BindingDB 50000092 |
| IC50 | 0.0043 uM | Not Available | BindingDB 50000092 |
| IC50 | 0.0085 uM | Not Available | BindingDB 50000092 |
References
- Chen X, Ji ZL, Chen YZ: TTD: Therapeutic Target Database. Nucleic Acids Res. 2002 Jan 1;30(1):412-5. [11752352 ]
- Yamada H, Shimoyama N, Sora I, Uhl GR, Fukuda Y, Moriya H, Shimoyama M: Morphine can produce analgesia via spinal kappa opioid receptors in the absence of mu opioid receptors. Brain Res. 2006 Apr 14;1083(1):61-9. Epub 2006 Mar 10. [16530171 ]
- Han W, Kasai S, Hata H, Takahashi T, Takamatsu Y, Yamamoto H, Uhl GR, Sora I, Ikeda K: Intracisternal A-particle element in the 3' noncoding region of the mu-opioid receptor gene in CXBK mice: a new genetic mechanism underlying differences in opioid sensitivity. Pharmacogenet Genomics. 2006 Jun;16(6):451-60. [16708053 ]
- Choi HS, Kim CS, Hwang CK, Song KY, Wang W, Qiu Y, Law PY, Wei LN, Loh HH: The opioid ligand binding of human mu-opioid receptor is modulated by novel splice variants of the receptor. Biochem Biophys Res Commun. 2006 May 19;343(4):1132-40. Epub 2006 Mar 23. [16580639 ]
- Castro RR, Cunha FQ, Silva FS Jr, Rocha FA: A quantitative approach to measure joint pain in experimental osteoarthritis--evidence of a role for nitric oxide. Osteoarthritis Cartilage. 2006 Aug;14(8):769-76. Epub 2006 Mar 31. [16580848 ]
- Johnson EA, Oldfield S, Braksator E, Gonzalez-Cuello A, Couch D, Hall KJ, Mundell SJ, Bailey CP, Kelly E, Henderson G: Agonist-selective mechanisms of mu-opioid receptor desensitization in human embryonic kidney 293 cells. Mol Pharmacol. 2006 Aug;70(2):676-85. Epub 2006 May 8. [16682505 ]
- Hanessian S, Parthasarathy S, Mauduit M, Payza K: The power of visual imagery in drug design. Isopavines as a new class of morphinomimetics and their human opioid receptor binding activity. J Med Chem. 2003 Jan 2;46(1):34-48. [12502358 ]
- Marastoni M, Salvadori S, Balboni G, Borea PA, Marzola G, Tomatis R: Synthesis and activity profiles of new dermorphin-(1-4) peptide analogues. J Med Chem. 1987 Sep;30(9):1538-42. [2887656 ]
- De Marco R, Tolomelli A, Spampinato S, Bedini A, Gentilucci L: Opioid activity profiles of oversimplified peptides lacking in the protonable N-terminus. J Med Chem. 2012 Nov 26;55(22):10292-6. doi: 10.1021/jm301213s. Epub 2012 Oct 19. [22995061 ]
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK: BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007 Jan;35(Database issue):D198-201. Epub 2006 Dec 1. [17145705 ]
- Varamini P, Mansfeld FM, Blanchfield JT, Wyse BD, Smith MT, Toth I: Synthesis and biological evaluation of an orally active glycosylated endomorphin-1. J Med Chem. 2012 Jun 28;55(12):5859-67. doi: 10.1021/jm300418d. Epub 2012 Jun 15. [22680612 ]
- Wentland MP, Duan W, Cohen DJ, Bidlack JM: Selective protection and functionalization of morphine: synthesis and opioid receptor binding properties of 3-amino-3-desoxymorphine derivatives. J Med Chem. 2000 Sep 21;43(19):3558-65. [11000010 ]
- Zhang A, Xiong W, Hilbert JE, DeVita EK, Bidlack JM, Neumeyer JL: 2-aminothiazole-derived opioids. Bioisosteric replacement of phenols. J Med Chem. 2004 Apr 8;47(8):1886-8. [15055988 ]
- Peng X, Knapp BI, Bidlack JM, Neumeyer JL: Synthesis and preliminary in vitro investigation of bivalent ligands containing homo- and heterodimeric pharmacophores at mu, delta, and kappa opioid receptors. J Med Chem. 2006 Jan 12;49(1):256-62. [16392810 ]
- Neumeyer JL, Zhang B, Zhang T, Sromek AW, Knapp BI, Cohen DJ, Bidlack JM: Synthesis, binding affinity, and functional in vitro activity of 3-benzylaminomorphinan and 3-benzylaminomorphine ligands at opioid receptors. J Med Chem. 2012 Apr 26;55(8):3878-90. doi: 10.1021/jm3001086. Epub 2012 Apr 4. [22439881 ]
- Nieland NP, Moynihan HA, Carrington S, Broadbear J, Woods JH, Traynor JR, Husbands SM, Lewis JW: Structural determinants of opioid activity in derivatives of 14-aminomorphinones: effect of substitution in the aromatic ring of cinnamoylaminomorphinones and codeinones. J Med Chem. 2006 Aug 24;49(17):5333-8. [16913723 ]
- Rennison D, Neal AP, Cami-Kobeci G, Aceto MD, Martinez-Bermejo F, Lewis JW, Husbands SM: Cinnamoyl derivatives of 7alpha-aminomethyl-6,14-endo-ethanotetrahydrothebaine and 7alpha-aminomethyl-6,14-endo-ethanotetrahydrooripavine and related opioid ligands. J Med Chem. 2007 Oct 18;50(21):5176-82. Epub 2007 Sep 22. [17887741 ]
- Halfpenny PR, Hill RG, Horwell DC, Hughes J, Hunter JC, Johnson S, Rees DC: Highly selective kappa-opioid analgesics. 2. Synthesis and structure-activity relationships of novel N-[(2-aminocyclohexyl)aryl]acetamide derivatives. J Med Chem. 1989 Jul;32(7):1620-6. [2567782 ]
- Clark CR, Halfpenny PR, Hill RG, Horwell DC, Hughes J, Jarvis TC, Rees DC, Schofield D: Highly selective kappa opioid analgesics. Synthesis and structure-activity relationships of novel N-[(2-aminocyclohexyl)aryl]acetamide and N-[(2-aminocyclohexyl)aryloxy]acetamide derivatives. J Med Chem. 1988 Apr;31(4):831-6. [2832603 ]
- Majumdar S, Burgman M, Haselton N, Grinnell S, Ocampo J, Pasternak AR, Pasternak GW: Generation of novel radiolabeled opiates through site-selective iodination. Bioorg Med Chem Lett. 2011 Jul 1;21(13):4001-4. doi: 10.1016/j.bmcl.2011.05.008. Epub 2011 May 8. [21621410 ]
- Zhang Y, Lee YS, Rothman RB, Dersch CM, Deschamps JR, Jacobson AE, Rice KC: Probes for narcotic receptor mediated phenomena. 39. Enantiomeric N-substituted benzofuro[2,3-c]pyridin-6-ols: synthesis and topological relationship to oxide-bridged phenylmorphans. J Med Chem. 2009 Dec 10;52(23):7570-9. doi: 10.1021/jm9004225. [19627147 ]
- Li F, Folk JE, Cheng K, Kurimura M, Deck JA, Deschamps JR, Rothman RB, Dersch CM, Jacobson AE, Rice KC: Probes for narcotic receptor mediated phenomena. 43. Synthesis of the ortho-a and para-a, and improved synthesis and optical resolution of the ortho-b and para-b oxide-bridged phenylmorphans: compounds with moderate to low opioid-receptor affinity. Bioorg Med Chem. 2011 Jul 15;19(14):4330-7. doi: 10.1016/j.bmc.2011.05.035. Epub 2011 May 24. [21684752 ]
- Kim JH, Deschamps JR, Rothman RB, Dersch CM, Folk JE, Cheng K, Jacobson AE, Rice KC: Probes for narcotic receptor mediated phenomena. Part 42: synthesis and in vitro pharmacological characterization of the N-methyl and N-phenethyl analogues of the racemic ortho-c and para-c oxide-bridged phenylmorphans. Bioorg Med Chem. 2011 Jun 1;19(11):3434-43. doi: 10.1016/j.bmc.2011.04.028. Epub 2011 Apr 22. [21570305 ]
- Iyer MR, Lee YS, Deschamps JR, Dersch CM, Rothman RB, Jacobson AE, Rice KC: Probes for narcotic receptor mediated phenomena. 44. Synthesis of an N-substituted 4-hydroxy-5-(3-hydroxyphenyl)morphan with high affinity and selective mu-antagonist activity. Eur J Med Chem. 2012 Apr;50:44-54. doi: 10.1016/j.ejmech.2012.01.025. Epub 2012 Jan 20. [22341895 ]
- Anzini M, Canullo L, Braile C, Cappelli A, Gallelli A, Vomero S, Menziani MC, De Benedetti PG, Rizzo M, Collina S, Azzolina O, Sbacchi M, Ghelardini C, Galeotti N: Synthesis, biological evaluation, and receptor docking simulations of 2-[(acylamino)ethyl]-1,4-benzodiazepines as kappa-opioid receptor agonists endowed with antinociceptive and antiamnesic activity. J Med Chem. 2003 Aug 28;46(18):3853-64. [12930147 ]
- Vecchietti V, Clarke GD, Colle R, Dondio G, Giardina G, Petrone G, Sbacchi M: Substituted 1-(aminomethyl)-2-(arylacetyl)-1,2,3,4-tetrahydroisoquinolines: a novel class of very potent antinociceptive agents with varying degrees of selectivity for kappa and mu opioid receptors. J Med Chem. 1992 Aug 7;35(16):2970-8. [1323679 ]
- Vecchietti V, Clarke GD, Colle R, Giardina G, Petrone G, Sbacchi M: (1S)-1-(aminomethyl)-2-(arylacetyl)-1,2,3,4-tetrahydroisoquinoline and heterocycle-condensed tetrahydropyridine derivatives: members of a novel class of very potent kappa opioid analgesics. J Med Chem. 1991 Aug;34(8):2624-33. [1652025 ]
- Vecchietti V, Giordani A, Giardina G, Colle R, Clarke GD: (2S)-1-(arylacetyl)-2-(aminomethyl)piperidine derivatives: novel, highly selective kappa opioid analgesics. J Med Chem. 1991 Jan;34(1):397-403. [1846921 ]
- Wang C, McFadyen IJ, Traynor JR, Mosberg HI: Design of a high affinity peptidomimetic opioid agonist from peptide pharmacophore models. Bioorg Med Chem Lett. 1998 Oct 6;8(19):2685-8. [9873603 ]
- Yoshino H, Nakazawa T, Arakawa Y, Kaneko T, Tsuchiya Y, Matsunaga M, Araki S, Ikeda M, Yamatsu K, Tachibana S: Synthesis and structure-activity relationships of dynorphin A-(1-8) amide analogues. J Med Chem. 1990 Jan;33(1):206-12. [1967312 ]
- Shreder K, Zhang L, Dang T, Yaksh TL, Umeno H, DeHaven R, Daubert J, Goodman M: Synthesis and biological activity of a novel methylamine-bridged enkephalin analogue (MABE): a new route to cyclic peptides and peptidomimetics. J Med Chem. 1998 Jul 2;41(14):2631-5. [9651168 ]
- Rew Y, Malkmus S, Svensson C, Yaksh TL, Chung NN, Schiller PW, Cassel JA, DeHaven RN, Taulane JP, Goodman M: Synthesis and biological activities of cyclic lanthionine enkephalin analogues: delta-opioid receptor selective ligands. J Med Chem. 2002 Aug 15;45(17):3746-54. [12166947 ]
- Tsai YC, Liou JP, Liao R, Cheng CY, Tao PL: C-alkylated spiro[benzofuran-3(2H),4'-1'-methyl-piperidine-7-ols] as potent opioids: a conformation-activity study. Bioorg Med Chem Lett. 1998 Jul 21;8(14):1813-8. [9873439 ]
- Tzschentke TM, Christoph T, Kogel B, Schiene K, Hennies HH, Englberger W, Haurand M, Jahnel U, Cremers TI, Friderichs E, De Vry J: (-)-(1R,2R)-3-(3-dimethylamino-1-ethyl-2-methyl-propyl)-phenol hydrochloride (tapentadol HCl): a novel mu-opioid receptor agonist/norepinephrine reuptake inhibitor with broad-spectrum analgesic properties. J Pharmacol Exp Ther. 2007 Oct;323(1):265-76. Epub 2007 Jul 26. [17656655 ]
- General Function:
- Opioid receptor activity
- Specific Function:
- G-protein coupled receptor that functions as receptor for endogenous enkephalins and for a subset of other opioids. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors, such as adenylate cyclase. Signaling leads to the inhibition of adenylate cyclase activity. Inhibits neurotransmitter release by reducing calcium ion currents and increasing potassium ion conductance. Plays a role in the perception of pain and in opiate-mediated analgesia. Plays a role in developing analgesic tolerance to morphine.
- Gene Name:
- OPRD1
- Uniprot ID:
- P41143
- Molecular Weight:
- 40368.235 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 0.1 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 0.14 uM | Not Available | BindingDB 50000092 |
| Inhibitory | 1 uM | Not Available | BindingDB 50000092 |
References
- Chen X, Ji ZL, Chen YZ: TTD: Therapeutic Target Database. Nucleic Acids Res. 2002 Jan 1;30(1):412-5. [11752352 ]
- Yamada H, Shimoyama N, Sora I, Uhl GR, Fukuda Y, Moriya H, Shimoyama M: Morphine can produce analgesia via spinal kappa opioid receptors in the absence of mu opioid receptors. Brain Res. 2006 Apr 14;1083(1):61-9. Epub 2006 Mar 10. [16530171 ]
- Dortch-Carnes J, Russell KR: Morphine-induced reduction of intraocular pressure and pupil diameter: role of nitric oxide. Pharmacology. 2006;77(1):17-24. Epub 2006 Mar 13. [16534251 ]
- Koch T, Wu DF, Yang LQ, Brandenburg LO, Hollt V: Role of phospholipase D2 in the agonist-induced and constitutive endocytosis of G-protein coupled receptors. J Neurochem. 2006 Apr;97(2):365-72. Epub 2006 Mar 15. [16539674 ]
- Galeotti N, Stefano GB, Guarna M, Bianchi E, Ghelardini C: Signaling pathway of morphine induced acute thermal hyperalgesia in mice. Pain. 2006 Aug;123(3):294-305. Epub 2006 May 2. [16650582 ]
- Asensio VJ, Miralles A, Garcia-Sevilla JA: Stimulation of mitogen-activated protein kinase kinases (MEK1/2) by mu-, delta- and kappa-opioid receptor agonists in the rat brain: regulation by chronic morphine and opioid withdrawal. Eur J Pharmacol. 2006 Jun 6;539(1-2):49-56. Epub 2006 Apr 6. [16678156 ]
- Standifer KM, Cheng J, Brooks AI, Honrado CP, Su W, Visconti LM, Biedler JL, Pasternak GW: Biochemical and pharmacological characterization of mu, delta and kappa 3 opioid receptors expressed in BE(2)-C neuroblastoma cells. J Pharmacol Exp Ther. 1994 Sep;270(3):1246-55. [7932177 ]
- Toll L, Berzetei-Gurske IP, Polgar WE, Brandt SR, Adapa ID, Rodriguez L, Schwartz RW, Haggart D, O'Brien A, White A, Kennedy JM, Craymer K, Farrington L, Auh JS: Standard binding and functional assays related to medications development division testing for potential cocaine and opiate narcotic treatment medications. NIDA Res Monogr. 1998 Mar;178:440-66. [9686407 ]
- Raynor K, Kong H, Chen Y, Yasuda K, Yu L, Bell GI, Reisine T: Pharmacological characterization of the cloned kappa-, delta-, and mu-opioid receptors. Mol Pharmacol. 1994 Feb;45(2):330-4. [8114680 ]
- General Function:
- Vitamin d3 25-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e.g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide (PubMed:11159812). Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro (PubMed:21576599).
- Gene Name:
- CYP3A4
- Uniprot ID:
- P08684
- Molecular Weight:
- 57342.67 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | >50 uM | Not Available | BindingDB 50000092 |
References
- Schwab D, Fischer H, Tabatabaei A, Poli S, Huwyler J: Comparison of in vitro P-glycoprotein screening assays: recommendations for their use in drug discovery. J Med Chem. 2003 Apr 24;46(9):1716-25. [12699389 ]
- General Function:
- Xenobiotic-transporting atpase activity
- Specific Function:
- Energy-dependent efflux pump responsible for decreased drug accumulation in multidrug-resistant cells.
- Gene Name:
- ABCB1
- Uniprot ID:
- P08183
- Molecular Weight:
- 141477.255 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | >50 uM | Not Available | BindingDB 50000092 |
References
- Schwab D, Fischer H, Tabatabaei A, Poli S, Huwyler J: Comparison of in vitro P-glycoprotein screening assays: recommendations for their use in drug discovery. J Med Chem. 2003 Apr 24;46(9):1716-25. [12699389 ]
- General Function:
- Nociceptin receptor activity
- Specific Function:
- G-protein coupled opioid receptor that functions as receptor for the endogenous neuropeptide nociceptin. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors. Signaling via G proteins mediates inhibition of adenylate cyclase activity and calcium channel activity. Arrestins modulate signaling via G proteins and mediate the activation of alternative signaling pathways that lead to the activation of MAP kinases. Plays a role in modulating nociception and the perception of pain. Plays a role in the regulation of locomotor activity by the neuropeptide nociceptin.
- Gene Name:
- OPRL1
- Uniprot ID:
- P41146
- Molecular Weight:
- 40692.775 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 0.0139 uM | Not Available | BindingDB 50000092 |
References
- Crooks PA, Kottayil SG, Al-Ghananeem AM, Byrn SR, Butterfield DA: Opiate receptor binding properties of morphine-, dihydromorphine-, and codeine 6-O-sulfate ester congeners. Bioorg Med Chem Lett. 2006 Aug 15;16(16):4291-5. Epub 2006 Jun 13. [16777416 ]
- General Function:
- Voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization
- Specific Function:
- Pore-forming (alpha) subunit of voltage-gated inwardly rectifying potassium channel. Channel properties are modulated by cAMP and subunit assembly. Mediates the rapidly activating component of the delayed rectifying potassium current in heart (IKr). Isoforms USO have no channel activity by themself, but modulates channel characteristics by forming heterotetramers with other isoforms which are retained intracellularly and undergo ubiquitin-dependent degradation.
- Gene Name:
- KCNH2
- Uniprot ID:
- Q12809
- Molecular Weight:
- 126653.52 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 1000 uM | Not Available | BindingDB 50000092 |
References
- Tobita M, Nishikawa T, Nagashima R: A discriminant model constructed by the support vector machine method for HERG potassium channel inhibitors. Bioorg Med Chem Lett. 2005 Jun 2;15(11):2886-90. [15911273 ]
- General Function:
- Opioid receptor activity
- Specific Function:
- Functions in lipid transport from the endoplasmic reticulum and is involved in a wide array of cellular functions probably through regulation of the biogenesis of lipid microdomains at the plasma membrane. Involved in the regulation of different receptors it plays a role in BDNF signaling and EGF signaling. Also regulates ion channels like the potassium channel and could modulate neurotransmitter release. Plays a role in calcium signaling through modulation together with ANK2 of the ITP3R-dependent calcium efflux at the endoplasmic reticulum. Plays a role in several other cell functions including proliferation, survival and death. Originally identified for its ability to bind various psychoactive drugs it is involved in learning processes, memory and mood alteration (PubMed:16472803, PubMed:9341151). Necessary for proper mitochondrial axonal transport in motor neurons, in particular the retrograde movement of mitochondria (By similarity).
- Gene Name:
- SIGMAR1
- Uniprot ID:
- Q99720
- Molecular Weight:
- 25127.52 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | >100 uM | Not Available | BindingDB 50000092 |
References
- Cheng CY, Hsin LW, Tsai MC, Schmidt WK, Smith C, Tam SW: Synthesis and opioid activity of 7-oxygenated 2,3,4,4a,5,6,7,7a-octahydro-1H-benzofuro[3,2-e]isoquinolin-9-ols. J Med Chem. 1994 Sep 16;37(19):3121-7. [7932535 ]
- General Function:
- Secondary active organic cation transmembrane transporter activity
- Specific Function:
- Translocates a broad array of organic cations with various structures and molecular weights including the model compounds 1-methyl-4-phenylpyridinium (MPP), tetraethylammonium (TEA), N-1-methylnicotinamide (NMN), 4-(4-(dimethylamino)styryl)-N-methylpyridinium (ASP), the endogenous compounds choline, guanidine, histamine, epinephrine, adrenaline, noradrenaline and dopamine, and the drugs quinine, and metformin. The transport of organic cations is inhibited by a broad array of compounds like tetramethylammonium (TMA), cocaine, lidocaine, NMDA receptor antagonists, atropine, prazosin, cimetidine, TEA and NMN, guanidine, cimetidine, choline, procainamide, quinine, tetrabutylammonium, and tetrapentylammonium. Translocates organic cations in an electrogenic and pH-independent manner. Translocates organic cations across the plasma membrane in both directions. Transports the polyamines spermine and spermidine. Transports pramipexole across the basolateral membrane of the proximal tubular epithelial cells. The choline transport is activated by MMTS. Regulated by various intracellular signaling pathways including inhibition by protein kinase A activation, and endogenously activation by the calmodulin complex, the calmodulin-dependent kinase II and LCK tyrosine kinase.
- Gene Name:
- SLC22A1
- Uniprot ID:
- O15245
- Molecular Weight:
- 61153.345 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 28 uM | Not Available | BindingDB 50000092 |
References
- Ahlin G, Karlsson J, Pedersen JM, Gustavsson L, Larsson R, Matsson P, Norinder U, Bergstrom CA, Artursson P: Structural requirements for drug inhibition of the liver specific human organic cation transport protein 1. J Med Chem. 2008 Oct 9;51(19):5932-42. doi: 10.1021/jm8003152. Epub 2008 Sep 13. [18788725 ]
- General Function:
- Transmembrane signaling receptor activity
- Specific Function:
- Cooperates with LY96 and CD14 to mediate the innate immune response to bacterial lipopolysaccharide (LPS). Acts via MYD88, TIRAP and TRAF6, leading to NF-kappa-B activation, cytokine secretion and the inflammatory response (PubMed:9237759, PubMed:10835634). Also involved in LPS-independent inflammatory responses triggered by free fatty acids, such as palmitate, and Ni(2+). Responses triggered by Ni(2+) require non-conserved histidines and are, therefore, species-specific (PubMed:20711192). In complex with TLR6, promotes sterile inflammation in monocytes/macrophages in response to oxidized low-density lipoprotein (oxLDL) or amyloid-beta 42. In this context, the initial signal is provided by oxLDL- or amyloid-beta 42-binding to CD36. This event induces the formation of a heterodimer of TLR4 and TLR6, which is rapidly internalized and triggers inflammatory response, leading to the NF-kappa-B-dependent production of CXCL1, CXCL2 and CCL9 cytokines, via MYD88 signaling pathway, and CCL5 cytokine, via TICAM1 signaling pathway, as well as IL1B secretion. Binds electronegative LDL (LDL(-)) and mediates the cytokine release induced by LDL(-) (PubMed:23880187).
- Gene Name:
- TLR4
- Uniprot ID:
- O00206
- Molecular Weight:
- 95679.19 Da
References
- Hutchinson MR, Lewis SS, Coats BD, Rezvani N, Zhang Y, Wieseler JL, Somogyi AA, Yin H, Maier SF, Rice KC, Watkins LR: Possible involvement of toll-like receptor 4/myeloid differentiation factor-2 activity of opioid inactive isomers causes spinal proinflammation and related behavioral consequences. Neuroscience. 2010 May 19;167(3):880-93. doi: 10.1016/j.neuroscience.2010.02.011. Epub 2010 Feb 21. [20178837 ]