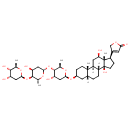You are using an unsupported browser. Please upgrade your browser to a newer version to get the best experience on Toxin, Toxin Target Database.
Digoxin (T3D2670)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2009-07-15 20:45:04 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:25:49 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D2670 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Digoxin | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Digoxin is a cardiac glycoside extracted from the foxglove plant, digitalis. It is widely used in the treatment of various heart conditions, namely atrial fibrillation, atrial flutter and congestive heart failure that cannot be controlled by other medication. Digoxin preparations are commonly marketed under the trade name Lanoxin. Digoxin has positive inotropic and negative chronotropic activity. It is used to control ventricular rate in atrial fibrillation and in the management of congestive heart failure with atrial fibrillation. Its use in congestive heart failure and sinus rhythm is less certain. The margin between toxic and therapeutic doses is small. (From Martindale, The Extra Pharmacopoeia, 30th ed, p666) Digoxin is a cardiotonic glycoside obtained mainly from Digitalis lanata; It consists of three sugars and the aglycone digoxigenin. Digoxin binds to a site on the extracellular aspect of the of the Na+/K+ ATPase pump in the membranes of heart cells (myocytes). This causes an increase in the level of sodium ions in the myocytes, which then leads to a rise in the level of calcium ions. The proposed mechanism is the following: inhibition of the Na+/K+ pump leads to increased Na+ levels, which in turn slows down the extrusion of Ca2+ via the Na+/Ca2+ exchange pump. Increased amounts of Ca2+ are then stored in the sarcoplasmic reticulum and released by each action potential, which is unchanged by digoxin. This is a different mechanism from that of catecholamines. Owing to its narrow therapeutic index (the margin between effectiveness and toxicity), side effects of digoxin are inevitable. Nausea, vomiting and GIT upset are common, especially in higher doses. Decreased conduction in the AV node can lead to AV blocks, increased intracellular Ca2+ causes a type of arrhythmia called bigeminy (coupled beats), eventually ventricular tachycardia or fibrillation. An often described but rarely seen side effect of digoxin is a disturbance of color vision (mostly yellow and green color) called xanthopsia. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C41H64O14 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 780.939 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 780.430 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 20830-75-5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 4-[(1S,2S,5S,7R,10R,11S,14R,15S,16R)-5-{[(2R,4S,5S,6R)-5-{[(2S,4S,5S,6R)-5-{[(2S,4S,5S,6R)-4,5-dihydroxy-6-methyloxan-2-yl]oxy}-4-hydroxy-6-methyloxan-2-yl]oxy}-4-hydroxy-6-methyloxan-2-yl]oxy}-11,16-dihydroxy-2,15-dimethyltetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]-2,5-dihydrofuran-2-one | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | digoxin | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [H][C@@]1(CC[C@]2(O)[C@]3([H])CC[C@]4([H])C[C@]([H])(CC[C@]4(C)[C@@]3([H])C[C@@]([H])(O)[C@]12C)O[C@@]1([H])C[C@]([H])(O)[C@]([H])(O[C@@]2([H])C[C@]([H])(O)[C@]([H])(O[C@@]3([H])C[C@]([H])(O)[C@]([H])(O)[C@@]([H])(C)O3)[C@@]([H])(C)O2)[C@@]([H])(C)O1)C1=CC(=O)OC1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C41H64O14/c1-19-36(47)28(42)15-34(50-19)54-38-21(3)52-35(17-30(38)44)55-37-20(2)51-33(16-29(37)43)53-24-8-10-39(4)23(13-24)6-7-26-27(39)14-31(45)40(5)25(9-11-41(26,40)48)22-12-32(46)49-18-22/h12,19-21,23-31,33-38,42-45,47-48H,6-11,13-18H2,1-5H3/t19-,20-,21-,23-,24+,25-,26-,27+,28+,29+,30+,31-,33+,34+,35+,36-,37-,38-,39+,40+,41+/m1/s1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=LTMHDMANZUZIPE-PUGKRICDSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as cardenolide glycosides and derivatives. Cardenolide glycosides and derivatives are compounds containing a carbohydrate glycosidically bound to the cardenolide moiety. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Steroids and steroid derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Steroid lactones | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Cardenolide glycosides and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteropolycyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | Colorless to white crystals; or white crystalline powder (1). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Injestion or dermal contact. (25) Absorption of digoxin from the elixir pediatric formulation has been demonstrated to be 70% to 85% complete (90% to 100% from the capsules, and 60% to 80% for tablets). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Digoxin binds to a site on the extracellular aspect of the alpha-subunit of the Na+/K+ ATPase pump in the membranes of heart cells (myocytes) and decreases its function. This causes an increase in the level of sodium ions in the myocytes. This effect causes an increase in the length the cardiac action potential, which when combined with the effects of digoxin on the parasympathetic nervous system, lead to a decrease in heart rate. Increased amounts of calcium are then stored in the sarcoplasmic reticulum and released by each action potential, which is unchanged by digoxin. This leads to increased contractility of the heart. Digoxin also increases vagal activity via its action on the central nervous system, thus decreasing the conduction of electrical impulses through the AV node. (23) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Hepatic (but not dependent upon the cytochrome P-450 system). The end metabolites, which include 3 b-digoxigenin, 3-keto-digoxigenin, and their glucuronide and sulfate conjugates, are polar in nature and are postulated to be formed via hydrolysis, oxidation, and conjugation. Route of Elimination: Following intravenous administration to healthy volunteers, 50% to 70% of a digoxin dose is excreted unchanged in the urine. Half Life: 3.5 to 5 days | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | LD50 = 7.8 mg/kg (orally in mice). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | 2B, possibly carcinogenic to humans. (26) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | For the treatment and management of congestive cardiac insufficiency, arrhythmias and heart failure. Digoxin is a plant toxin found in the foxglove plant (Digitalis lanata). It is used as a drug to treat various heart conditions, namely atrial fibrillation, atrial flutter and sometimes heart failure. (23) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Digoxin mainly affects the heart. (23) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Adverse affects of digoxin include loss of appetite, nausea, vomiting, diarrhea, blurred vision, visual disturbances (yellow-green halos), confusion, drowsiness, dizziness, nightmares, agitation, and/or depression, as well as a higher acute sense of sensual activities. (23) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Treatment of dioxin overdose includes supportive measure and administration of the antidote, antidigoxin (DIGIBIND). Toxicity can also be treated with higher than normal doses of potassium. (23) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | DB00390 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB01917 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 2724385 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL1751 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 2006532 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C06956 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | 108950 , 177720 , 309801 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 4551 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Digoxin | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | DGX | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Digoxin | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Wolfgang Voigtlander, Fritz Kaiser, Wolfgang Schaumann, Kurt Stach, “Preparation of C22-alkyl derivative of digoxin.” U.S. Patent US3981862, issued October, 1972. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Link | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Steroid hormone binding
- Specific Function:
- This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients.
- Gene Name:
- ATP1A1
- Uniprot ID:
- P05023
- Molecular Weight:
- 112895.01 Da
References
- Ravikumar A, Arun P, Devi KV, Augustine J, Kurup PA: Isoprenoid pathway and free radical generation and damage in neuropsychiatric disorders. Indian J Exp Biol. 2000 May;38(5):438-46. [11272406 ]
- Chen JJ, Wang PS, Chien EJ, Wang SW: Direct inhibitory effect of digitalis on progesterone release from rat granulosa cells. Br J Pharmacol. 2001 Apr;132(8):1761-8. [11309248 ]
- Ke YS, Liu ZF, Yang H, Yang T, Huang JS, Rui SB, Cheng GH, Wang YX: Effect of anti-digoxin antiserum on endoxin and membrane ATPase activity in hypoxia-reoxygenation induced myocardial injury. Acta Pharmacol Sin. 2000 Apr;21(4):345-7. [11324464 ]
- Kumar AR, Kurup PA: A hypothalamic digoxin mediated model for conscious and subliminal perception. J Neural Transm. 2001;108(7):855-68. [11515751 ]
- Aizman O, Uhlen P, Lal M, Brismar H, Aperia A: Ouabain, a steroid hormone that signals with slow calcium oscillations. Proc Natl Acad Sci U S A. 2001 Nov 6;98(23):13420-4. Epub 2001 Oct 30. [11687608 ]
- Wikipedia. Digoxin. Last Updated 8 July 2009. [Link]
- General Function:
- Sodium-independent organic anion transmembrane transporter activity
- Specific Function:
- Mediates the Na(+)-independent uptake of organic anions such as pravastatin, taurocholate, methotrexate, dehydroepiandrosterone sulfate, 17-beta-glucuronosyl estradiol, estrone sulfate, prostaglandin E2, thromboxane B2, leukotriene C3, leukotriene E4, thyroxine and triiodothyronine. Involved in the clearance of bile acids and organic anions from the liver.
- Gene Name:
- SLCO1B1
- Uniprot ID:
- Q9Y6L6
- Molecular Weight:
- 76447.99 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 3.236 uM | Not Available | BindingDB 46355 |
References
- Dagenais C, Ducharme J, Pollack GM: Uptake and efflux of the peptidic delta-opioid receptor agonist. Neurosci Lett. 2001 Apr 6;301(3):155-8. [11257421 ]
- Sugiyama D, Kusuhara H, Shitara Y, Abe T, Meier PJ, Sekine T, Endou H, Suzuki H, Sugiyama Y: Characterization of the efflux transport of 17beta-estradiol-D-17beta-glucuronide from the brain across the blood-brain barrier. J Pharmacol Exp Ther. 2001 Jul;298(1):316-22. [11408557 ]
- Hagenbuch N, Reichel C, Stieger B, Cattori V, Fattinger KE, Landmann L, Meier PJ, Kullak-Ublick GA: Effect of phenobarbital on the expression of bile salt and organic anion transporters of rat liver. J Hepatol. 2001 Jun;34(6):881-7. [11451172 ]
- Gao B, Wenzel A, Grimm C, Vavricka SR, Benke D, Meier PJ, Reme CE: Localization of organic anion transport protein 2 in the apical region of rat retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2002 Feb;43(2):510-4. [11818398 ]
- Shitara Y, Sugiyama D, Kusuhara H, Kato Y, Abe T, Meier PJ, Itoh T, Sugiyama Y: Comparative inhibitory effects of different compounds on rat oatpl (slc21a1)- and Oatp2 (Slc21a5)-mediated transport. Pharm Res. 2002 Feb;19(2):147-53. [11883641 ]
- De Bruyn T, van Westen GJ, Ijzerman AP, Stieger B, de Witte P, Augustijns PF, Annaert PP: Structure-based identification of OATP1B1/3 inhibitors. Mol Pharmacol. 2013 Jun;83(6):1257-67. doi: 10.1124/mol.112.084152. Epub 2013 Apr 9. [23571415 ]
- General Function:
- Vitamin d3 25-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e.g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide (PubMed:11159812). Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro (PubMed:21576599).
- Gene Name:
- CYP3A4
- Uniprot ID:
- P08684
- Molecular Weight:
- 57342.67 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | >50 uM | Not Available | BindingDB 46355 |
References
- Schwab D, Fischer H, Tabatabaei A, Poli S, Huwyler J: Comparison of in vitro P-glycoprotein screening assays: recommendations for their use in drug discovery. J Med Chem. 2003 Apr 24;46(9):1716-25. [12699389 ]
- General Function:
- Xenobiotic-transporting atpase activity
- Specific Function:
- Energy-dependent efflux pump responsible for decreased drug accumulation in multidrug-resistant cells.
- Gene Name:
- ABCB1
- Uniprot ID:
- P08183
- Molecular Weight:
- 141477.255 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | >50 uM | Not Available | BindingDB 46355 |
References
- Schwab D, Fischer H, Tabatabaei A, Poli S, Huwyler J: Comparison of in vitro P-glycoprotein screening assays: recommendations for their use in drug discovery. J Med Chem. 2003 Apr 24;46(9):1716-25. [12699389 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear receptor that binds DNA as a monomer to ROR response elements (RORE) containing a single core motif half-site 5'-AGGTCA-3' preceded by a short A-T-rich sequence. Key regulator of cellular differentiation, immunity, peripheral circadian rhythm as well as lipid, steroid, xenobiotics and glucose metabolism. Considered to have intrinsic transcriptional activity, have some natural ligands like oxysterols that act as agonists (25-hydroxycholesterol) or inverse agonists (7-oxygenated sterols), enhancing or repressing the transcriptional activity, respectively. Recruits distinct combinations of cofactors to target gene regulatory regions to modulate their transcriptional expression, depending on the tissue, time and promoter contexts. Regulates the circadian expression of clock genes such as CRY1, ARNTL/BMAL1 and NR1D1 in peripheral tissues and in a tissue-selective manner. Competes with NR1D1 for binding to their shared DNA response element on some clock genes such as ARNTL/BMAL1, CRY1 and NR1D1 itself, resulting in NR1D1-mediated repression or RORC-mediated activation of the expression, leading to the circadian pattern of clock genes expression. Therefore influences the period length and stability of the clock. Involved in the regulation of the rhythmic expression of genes involved in glucose and lipid metabolism, including PLIN2 and AVPR1A. Negative regulator of adipocyte differentiation through the regulation of early phase genes expression, such as MMP3. Controls adipogenesis as well as adipocyte size and modulates insulin sensitivity in obesity. In liver, has specific and redundant functions with RORA as positive or negative modulator of expression of genes encoding phase I and Phase II proteins involved in the metabolism of lipids, steroids and xenobiotics, such as SULT1E1. Also plays also a role in the regulation of hepatocyte glucose metabolism through the regulation of G6PC and PCK1. Regulates the rhythmic expression of PROX1 and promotes its nuclear localization (By similarity). Plays an indispensable role in the induction of IFN-gamma dependent anti-mycobacterial systemic immunity (PubMed:26160376).Isoform 2: Essential for thymopoiesis and the development of several secondary lymphoid tissues, including lymph nodes and Peyer's patches. Required for the generation of LTi (lymphoid tissue inducer) cells. Regulates thymocyte survival through DNA-binding on ROREs of target gene promoter regions and recruitment of coactivaros via the AF-2. Also plays a key role, downstream of IL6 and TGFB and synergistically with RORA, for lineage specification of uncommitted CD4(+) T-helper (T(H)) cells into T(H)17 cells, antagonizing the T(H)1 program. Probably regulates IL17 and IL17F expression on T(H) by binding to the essential enhancer conserved non-coding sequence 2 (CNS2) in the IL17-IL17F locus. May also play a role in the pre-TCR activation cascade leading to the maturation of alpha/beta T-cells and may participate in the regulation of DNA accessibility in the TCR-J(alpha) locus.
- Gene Name:
- RORC
- Uniprot ID:
- P51449
- Molecular Weight:
- 58194.845 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 2 uM | Not Available | BindingDB 46355 |
References
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK: BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007 Jan;35(Database issue):D198-201. Epub 2006 Dec 1. [17145705 ]
- General Function:
- Tumor necrosis factor receptor binding
- Specific Function:
- Signal transducer and transcription activator that mediates cellular responses to interferons (IFNs), cytokine KITLG/SCF and other cytokines and other growth factors. Following type I IFN (IFN-alpha and IFN-beta) binding to cell surface receptors, signaling via protein kinases leads to activation of Jak kinases (TYK2 and JAK1) and to tyrosine phosphorylation of STAT1 and STAT2. The phosphorylated STATs dimerize and associate with ISGF3G/IRF-9 to form a complex termed ISGF3 transcription factor, that enters the nucleus. ISGF3 binds to the IFN stimulated response element (ISRE) to activate the transcription of IFN-stimulated genes (ISG), which drive the cell in an antiviral state. In response to type II IFN (IFN-gamma), STAT1 is tyrosine- and serine-phosphorylated. It then forms a homodimer termed IFN-gamma-activated factor (GAF), migrates into the nucleus and binds to the IFN gamma activated sequence (GAS) to drive the expression of the target genes, inducing a cellular antiviral state. Becomes activated in response to KITLG/SCF and KIT signaling. May mediate cellular responses to activated FGFR1, FGFR2, FGFR3 and FGFR4.
- Gene Name:
- STAT1
- Uniprot ID:
- P42224
- Molecular Weight:
- 87334.175 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | >55.70 uM | Not Available | BindingDB 46355 |
References
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK: BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007 Jan;35(Database issue):D198-201. Epub 2006 Dec 1. [17145705 ]
- General Function:
- Sodium-independent organic anion transmembrane transporter activity
- Specific Function:
- Mediates the Na(+)-independent uptake of organic anions such as 17-beta-glucuronosyl estradiol, taurocholate, triiodothyronine (T3), leukotriene C4, dehydroepiandrosterone sulfate (DHEAS), methotrexate and sulfobromophthalein (BSP). Involved in the clearance of bile acids and organic anions from the liver.
- Gene Name:
- SLCO1B3
- Uniprot ID:
- Q9NPD5
- Molecular Weight:
- 77402.175 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 13.183 uM | Not Available | BindingDB 46355 |
References
- De Bruyn T, van Westen GJ, Ijzerman AP, Stieger B, de Witte P, Augustijns PF, Annaert PP: Structure-based identification of OATP1B1/3 inhibitors. Mol Pharmacol. 2013 Jun;83(6):1257-67. doi: 10.1124/mol.112.084152. Epub 2013 Apr 9. [23571415 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Steroid hormone receptors are ligand-activated transcription factors that regulate eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Transcription factor activity is modulated by bound coactivator and corepressor proteins. Transcription activation is down-regulated by NR0B2. Activated, but not phosphorylated, by HIPK3 and ZIPK/DAPK3.
- Gene Name:
- AR
- Uniprot ID:
- P10275
- Molecular Weight:
- 98987.9 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.03 uM | Tox21_AR_LUC_MDAKB2_Agonist | Tox21/NCGC |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear receptor that binds peroxisome proliferators such as hypolipidemic drugs and fatty acids. Once activated by a ligand, the nuclear receptor binds to DNA specific PPAR response elements (PPRE) and modulates the transcription of its target genes, such as acyl-CoA oxidase. It therefore controls the peroxisomal beta-oxidation pathway of fatty acids. Key regulator of adipocyte differentiation and glucose homeostasis. ARF6 acts as a key regulator of the tissue-specific adipocyte P2 (aP2) enhancer. Acts as a critical regulator of gut homeostasis by suppressing NF-kappa-B-mediated proinflammatory responses. Plays a role in the regulation of cardiovascular circadian rhythms by regulating the transcription of ARNTL/BMAL1 in the blood vessels (By similarity).
- Gene Name:
- PPARG
- Uniprot ID:
- P37231
- Molecular Weight:
- 57619.58 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.06 uM | ATG_PPARg_TRANS | Attagene |
| AC50 | 0.04 uM | Tox21_PPARg_BLA_Agonist_ratio | Tox21/NCGC |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear hormone receptor. The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Ligand-dependent nuclear transactivation involves either direct homodimer binding to a palindromic estrogen response element (ERE) sequence or association with other DNA-binding transcription factors, such as AP-1/c-Jun, c-Fos, ATF-2, Sp1 and Sp3, to mediate ERE-independent signaling. Ligand binding induces a conformational change allowing subsequent or combinatorial association with multiprotein coactivator complexes through LXXLL motifs of their respective components. Mutual transrepression occurs between the estrogen receptor (ER) and NF-kappa-B in a cell-type specific manner. Decreases NF-kappa-B DNA-binding activity and inhibits NF-kappa-B-mediated transcription from the IL6 promoter and displace RELA/p65 and associated coregulators from the promoter. Recruited to the NF-kappa-B response element of the CCL2 and IL8 promoters and can displace CREBBP. Present with NF-kappa-B components RELA/p65 and NFKB1/p50 on ERE sequences. Can also act synergistically with NF-kappa-B to activate transcription involving respective recruitment adjacent response elements; the function involves CREBBP. Can activate the transcriptional activity of TFF1. Also mediates membrane-initiated estrogen signaling involving various kinase cascades. Isoform 3 is involved in activation of NOS3 and endothelial nitric oxide production. Isoforms lacking one or several functional domains are thought to modulate transcriptional activity by competitive ligand or DNA binding and/or heterodimerization with the full length receptor. Essential for MTA1-mediated transcriptional regulation of BRCA1 and BCAS3. Isoform 3 can bind to ERE and inhibit isoform 1.
- Gene Name:
- ESR1
- Uniprot ID:
- P03372
- Molecular Weight:
- 66215.45 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.07 uM | Tox21_ERa_LUC_BG1_Agonist | Tox21/NCGC |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear receptor that binds DNA as a monomer to ROR response elements (RORE) containing a single core motif half-site 5'-AGGTCA-3' preceded by a short A-T-rich sequence. Considered to have intrinsic transcriptional activity, have some natural ligands such as all-trans retinoic acid (ATRA) and other retinoids which act as inverse agonists repressing the transcriptional activity. Required for normal postnatal development of rod and cone photoreceptor cells. Modulates rod photoreceptors differentiation at least by inducing the transcription factor NRL-mediated pathway. In cone photoreceptor cells, regulates transcription of OPN1SW. Involved in the regulation of the period length and stability of the circadian rhythm. May control cytoarchitectural patterning of neocortical neurons during development. May act in a dose-dependent manner to regulate barrel formation upon innervation of layer IV neurons by thalamocortical axons. May play a role in the suppression of osteoblastic differentiation through the inhibition of RUNX2 transcriptional activity (By similarity).Isoform 1 is critical for hindlimb motor control and for the differentiation of amacrine and horizontal cells in the retina. Regulates the expression of PTF1A synergistically with FOXN4 (By similarity).
- Gene Name:
- RORB
- Uniprot ID:
- Q92753
- Molecular Weight:
- 53219.385 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.07 uM | ATG_RORb_TRANS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Ligand-activated transcription factor. Key regulator of lipid metabolism. Activated by the endogenous ligand 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (16:0/18:1-GPC). Activated by oleylethanolamide, a naturally occurring lipid that regulates satiety. Receptor for peroxisome proliferators such as hypolipidemic drugs and fatty acids. Regulates the peroxisomal beta-oxidation pathway of fatty acids. Functions as transcription activator for the ACOX1 and P450 genes. Transactivation activity requires heterodimerization with RXRA and is antagonized by NR2C2. May be required for the propagation of clock information to metabolic pathways regulated by PER2.
- Gene Name:
- PPARA
- Uniprot ID:
- Q07869
- Molecular Weight:
- 52224.595 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.07 uM | ATG_PPARa_TRANS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transcriptional activator activity, rna polymerase ii core promoter proximal region sequence-specific binding
- Specific Function:
- Transcriptional regulator. Recognizes and binds to the DNA sequence 5'-CGCCCCCGC-3'(EGR-site). Activates the transcription of target genes whose products are required for mitogenesis and differentiation.
- Gene Name:
- EGR1
- Uniprot ID:
- P18146
- Molecular Weight:
- 57506.17 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.29 uM | ATG_EGR_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Oxygen binding
- Specific Function:
- Catalyzes the formation of aromatic C18 estrogens from C19 androgens.
- Gene Name:
- CYP19A1
- Uniprot ID:
- P11511
- Molecular Weight:
- 57882.48 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.32 uM | Tox21_Aromatase_Inhibition | Tox21/NCGC |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transcription regulatory region sequence-specific dna binding
- Specific Function:
- Transcription factor that binds to the octamer motif (5'-ATTTGCAT-3') and activates the promoters of the genes for some small nuclear RNAs (snRNA) and of genes such as those for histone H2B and immunoglobulins. Modulates transcription transactivation by NR3C1, AR and PGR (By similarity). In case of human herpes simplex virus (HSV) infection, POU2F1 forms a multiprotein-DNA complex with the viral transactivator protein VP16 and HCFC1 thereby enabling the transcription of the viral immediate early genes.
- Gene Name:
- POU2F1
- Uniprot ID:
- P14859
- Molecular Weight:
- 76470.82 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.39 uM | ATG_Oct_MLP_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- Functions as a transcription factor during endoplasmic reticulum (ER) stress by regulating the unfolded protein response (UPR). Required for cardiac myogenesis and hepatogenesis during embryonic development, and the development of secretory tissues such as exocrine pancreas and salivary gland (By similarity). Involved in terminal differentiation of B lymphocytes to plasma cells and production of immunoglobulins (PubMed:11460154). Modulates the cellular response to ER stress in a PIK3R-dependent manner (PubMed:20348923). Binds to the cis-acting X box present in the promoter regions of major histocompatibility complex class II genes (PubMed:8349596). Involved in VEGF-induced endothelial cell (EC) proliferation and retinal blood vessel formation during embryonic development but also for angiogenesis in adult tissues under ischemic conditions. Functions also as a major regulator of the UPR in obesity-induced insulin resistance and type 2 diabetes for the management of obesity and diabetes prevention (By similarity).Isoform 1: plays a role in the unconventional cytoplasmic splicing processing of its own mRNA triggered by the endoplasmic reticulum (ER) transmembrane endoribonuclease ENR1: upon ER stress, the emerging XBP1 polypeptide chain, as part of a mRNA-ribosome-nascent chain (R-RNC) complex, cotranslationally recruits its own unprocessed mRNA through transient docking to the ER membrane and translational pausing, therefore facilitating efficient IRE1-mediated XBP1 mRNA isoform 2 production (PubMed:19394296, PubMed:21233347). In endothelial cells (EC), associated with KDR, promotes IRE1-mediated XBP1 mRNA isoform 2 productions in a vascular endothelial growth factor (VEGF)-dependent manner, leading to EC proliferation and angiogenesis (PubMed:23529610). Functions as a negative feed-back regulator of the potent transcription factor XBP1 isoform 2 protein levels through proteasome-mediated degradation, thus preventing the constitutive activation of the ER stress response signaling pathway (PubMed:16461360, PubMed:25239945). Inhibits the transactivation activity of XBP1 isoform 2 in myeloma cells (By similarity). Acts as a weak transcriptional factor (PubMed:8657566). Together with HDAC3, contributes to the activation of NFE2L2-mediated HMOX1 transcription factor gene expression in a PI(3)K/mTORC2/Akt-dependent signaling pathway leading to EC survival under disturbed flow/oxidative stress (PubMed:25190803). Binds to the ER stress response element (ERSE) upon ER stress (PubMed:11779464). Binds to the consensus 5'-GATGACGTG[TG]N(3)[AT]T-3' sequence related to cAMP responsive element (CRE)-like sequences (PubMed:8657566). Binds the Tax-responsive element (TRE) present in the long terminal repeat (LTR) of T cell leukemia virus type 1 (HTLV-I) and to the TPA response elements (TRE) (PubMed:2321018, PubMed:2196176, PubMed:1903538, PubMed:8657566). Associates preferentially to the HDAC3 gene promoter region in a static flow-dependent manner (PubMed:25190803). Binds to the CDH5/VE-cadherin gene promoter region (PubMed:19416856).Isoform 2: functions as a stress-inducible potent transcriptional activator during endoplasmic reticulum (ER) stress by inducing unfolded protein response (UPR) target genes via binding to the UPR element (UPRE). Up-regulates target genes encoding ER chaperones and ER-associated degradation (ERAD) components to enhance the capacity of productive folding and degradation mechanism, respectively, in order to maintain the homeostasis of the ER under ER stress (PubMed:11779464, PubMed:25239945). Plays a role in the production of immunoglobulins and interleukin-6 in the presence of stimuli required for plasma cell differentiation (By similarity). Induces phospholipid biosynthesis and ER expansion (PubMed:15466483). Contributes to the VEGF-induced endothelial cell (EC) growth and proliferation in a Akt/GSK-dependent and/or -independent signaling pathway, respectively, leading to beta-catenin nuclear translocation and E2F2 gene expression (PubMed:23529610). Promotes umbilical vein EC apoptosis and atherosclerotisis development in a caspase-dependent signaling pathway, and contributes to VEGF-induced EC proliferation and angiogenesis in adult tissues under ischemic conditions (PubMed:19416856, PubMed:23529610). Involved in the regulation of endostatin-induced autophagy in EC through BECN1 transcriptional activation (PubMed:23184933). Plays a role as an oncogene by promoting tumor progression: stimulates zinc finger protein SNAI1 transcription to induce epithelial-to-mesenchymal (EMT) transition, cell migration and invasion of breast cancer cells (PubMed:25280941). Involved in adipocyte differentiation by regulating lipogenic gene expression during lactation. Plays a role in the survival of both dopaminergic neurons of the substantia nigra pars compacta (SNpc), by maintaining protein homeostasis and of myeloma cells. Increases insulin sensitivity in the liver as a response to a high carbohydrate diet, resulting in improved glucose tolerance. Improves also glucose homeostasis in an ER stress- and/or insulin-independent manner through both binding and proteasome-induced degradation of the transcription factor FOXO1, hence resulting in suppression of gluconeogenic genes expression and in a reduction of blood glucose levels. Controls the induction of de novo fatty acid synthesis in hepatocytes by regulating the expression of a subset of lipogenic genes in an ER stress- and UPR-independent manner (By similarity). Associates preferentially to the HDAC3 gene promoter region in a disturbed flow-dependent manner (PubMed:25190803). Binds to the BECN1 gene promoter region (PubMed:23184933). Binds to the CDH5/VE-cadherin gene promoter region (PubMed:19416856). Binds to the ER stress response element (ERSE) upon ER stress (PubMed:11779464). Binds to the 5'-CCACG-3' motif in the PPARG promoter (By similarity).
- Gene Name:
- XBP1
- Uniprot ID:
- P17861
- Molecular Weight:
- 28694.66 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.41 uM | ATG_Xbp1_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear hormone receptor. Transcription factor that mediates the action of vitamin D3 by controlling the expression of hormone sensitive genes. Recruited to promoters via its interaction with BAZ1B/WSTF which mediates the interaction with acetylated histones, an essential step for VDR-promoter association. Plays a central role in calcium homeostasis.
- Gene Name:
- VDR
- Uniprot ID:
- P11473
- Molecular Weight:
- 48288.64 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.52 uM | ATG_VDRE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]